In my recent
paper on central complex processing during flight, I used
BrainAligner to register z-stacks of Drosophila brains to one another. Getting it to work took a little fiddling, which I'll record below.
To install, follow directions
here.
Next, right-click brainaligner_linux_redhat_fedora_64bit and choose properties. Under the Permissions tab, click the "Allow executing file as program" checkbox.
In terminal, type
gedit ~/.bashrc
and add
export PATH="/home/$USER/<rest of path to directory>:$PATH"
at the end of the file to add <rest of path to directory> to your path.
It gave me the following error
"error while loading shared libraries: libtiff.so.3: cannot open shared object file: No such file or directory"
Based on
this stackoverflow answer, I ran
sudo ln -s /usr/lib/x86_64-linux-gnu/libtiff.so.4 /usr/lib/x86_64-linux-gnu/libtiff.so.3
to get it to recognize libtiff.so.3
And it seems to work!
To work with multiple color channels, you have to go to image>type>RGB color to change a composite image to a 3 channel color image. BrainAligner will assume the first channel is the reference channel to use in the warping, unless you give it a different channel number as an argument.
After much testing, I found that the following command worked well:
./brainaligner_linux_redhat_fedora_64bit -t ./20150101/Z3_8b_c_b3.tif -s ./20150101/Z1_8b_c_b3.tif -o ./20150101/warpedZ1.tif -w 10 -B 341 -x 1 -z 1 -X 1 -Z 1
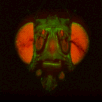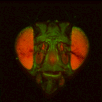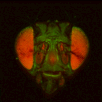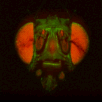
The file names of the image stacks remind me that after converting each z-stack to RGB color (image>type/RGB color in imageJ), I binned every three pixels. This resulted in stacks that were 341x341 pixels, and the x-y resolution was equal to the z resolution. BrainAligner worked in about 2 hours.
Definitely check the warped stack afterwards, because BrainAligner will fail if the initial two stacks are not very similar to begin with. (I was aligning two stack from the same animal before and after photoactivation, which was a relatively easy alignment problem.)